Before I start this post I would just like to state that the views expressed below are my own and not that of my employer.
For the last 18 months I've been trying to buy an ELISA plate reader, with no success.
Admittedly enzyme-linked immunosorbent assays (ELISA) are not new and have in fact been around for at least 30 years (I first heard about them during my first degree, and they were viewed as a replacement for the radio-immuno assay (RIA)). So, on the one hand you could be argued that the technology is old and well-established, or that it's a technology that is had its day. Either way you would expect the market to be mature and able to provide suitable equipment.
When I started looking for an ELISA plate reader I was struck by how old all the machines appeared to be (in a number of cases designed in the 1990s, and a few machines in the early to mid-2000s), the poor design of their user interface and how difficult it seemed to be to get data in and out of the machine (one company offered me a machine with an attached dot-matrix printer - I had no idea such printers were still made, can you even get the paper). What was really surprisingly was how few of the machines could be networked, and how old the input/output ports were on the machines.
What I was looking for was a modern machine that could easily export results either directly to a server, or to a USB data stick. You wouldn't think it would be that difficult to find such a machine? How wrong I was....
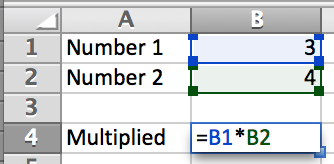
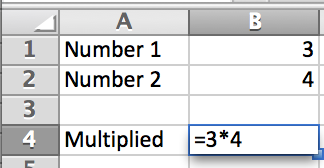
After two rounds of procurement (nothing suitable in the first round, so I dropped my requirements) I finally settled on the HumaReader HS as it appeared to be the best of a bad bunch. The machine, from what I could see in the literature, did have a rather clunky user interface but, and this was in my opinion it's big selling point, it did have two USB ports, a LAN port, and an SD card reader. From what I could gather from various promotional leaflets and the user manual, the machine was able to export data and methods to the USB ports, and it could be connected to a computer for direct export to Excel (see below). To me this seemed to be a good match as it meant we could easily collect and export data from 25 to 30 ELISA plates in an undergraduate practical and then redistribute the data to the students for analysis.

The HumaReader HS ELISA plate reader - it can read plates, but you can't get the data out of it...
The machine was delivered to us after 8 to 10 weeks and installed. It was only at this point it became apparent that the export to the USB stick was in a proprietary format (Why didn't the engineers think to do a simple CSV export? I guess they were not very good engineers?) that could only be read by another HumaReader HS machine. I really cannot see a reason for wanting to do this? Maybe the methods when setting up a number of machines, but why the data?

Ports on the back of the HumaReader HS ELISA plate reader - there may be data ports, but don't make the mistake I did and actually think you can export data
So, strike one, no USB export. So to my fallback plane... Export to Excel.
Unfortunately the HumaReader HS doesn't even seem to be able to export to Excel. When I raised this with the company (both the local supplier and the actual maker of the machine) I didn't receive a satisfactory response. In fact today I received the email below:
"Good day to you.
After several attempts, we managed to sort the data to excel format by using a customized Microsoft word Macros’ script.
The process is involved few steps as following:
1) Transfer the readings from ELISA reader to Laptop (LIS format).
2) Open the data which in LIS format to notepad, copy and paste the data to Microsoft word.
3) Sort the data by using a customized macros’ script and the data will be saved in excel format.
4) Open the excel format data and sort the data accordingly.
In
Upon the sorting, the data is as following and for detail please refer to attach file."
Is the company serious? It would appear they are! To have to do this for 25 to 30 plates in an undergraduate practical would be an absolute nightmare. What century are we living in?

An extract from the HumaReader HS Reader promotional material...
How do other people cope? How do they get data out of the machine? Why would you design a machine from which you cannot export the data? Absolute madness.
Today I have told the company they can collect the HumaReader HS Reader from the lab, and refund the money.
So, I'm back in the market for an ELISA plate reader. As for the ELISA practical running in a few weeks? Guess I will have to be creative...
If you would like to support my blogging efforts, then please feel free to buy me a coffee at https://www.buymeacoffee.com/drnickm
Declared conflict of interest: I am really interested and supportive of open data standards that allow the sharing of data between scientists and different labs. I heave worked on data standards in proteomics and was involved in establishing the 'Minimum information about a proteomics experiment (MIAPE) standard.

 'Anaesthetised' flies
'Anaesthetised' flies