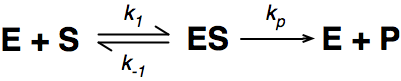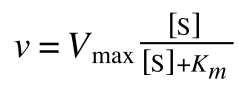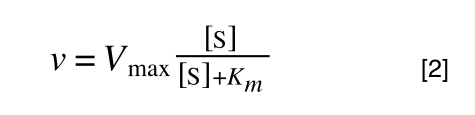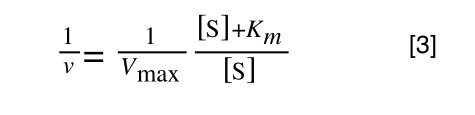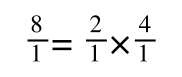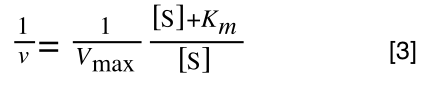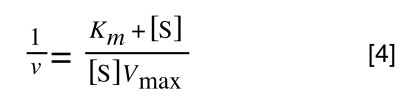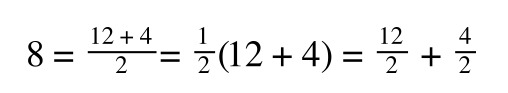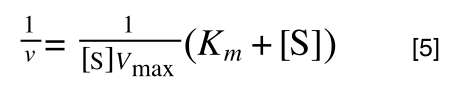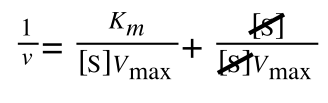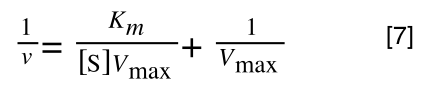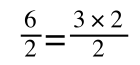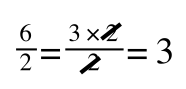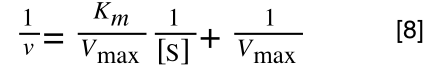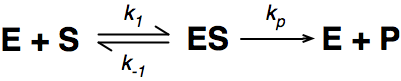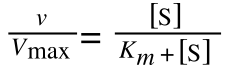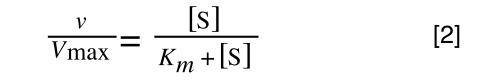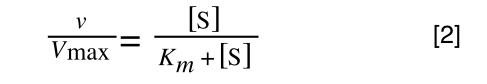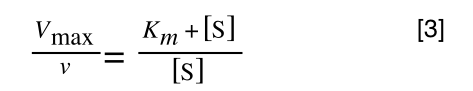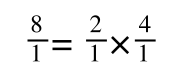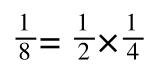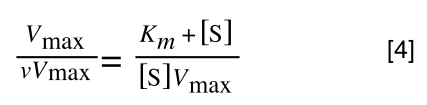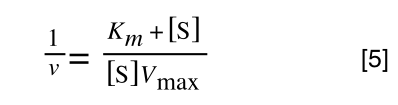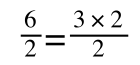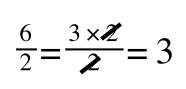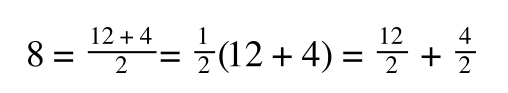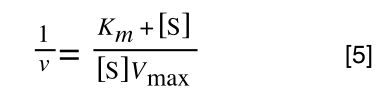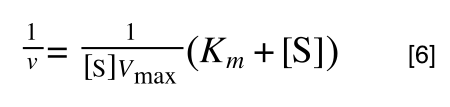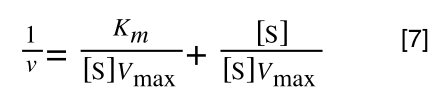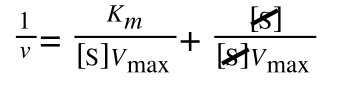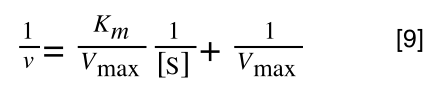OK, well, here is another way Deriving Lineweaver-Burk Reciprocal Plot from Michaelis Menten Equation, this time starting from a different point (the other approach is
here)...
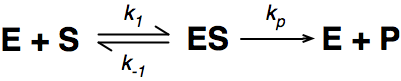
Where E = enzyme; S = substrate; ES = Enzyme substrate complex; and P = product
The Michaelis-Menton Equation given in your lectures describing the reaction is:
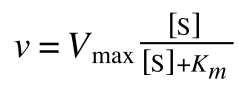
Where v = rate (initial velocity); Vmax = maximum velocity (100% of enzyme catalytic sites occupied); Km = Michaelis constant (concentration of substrate to achieve half Vmax); S = substrate concentration
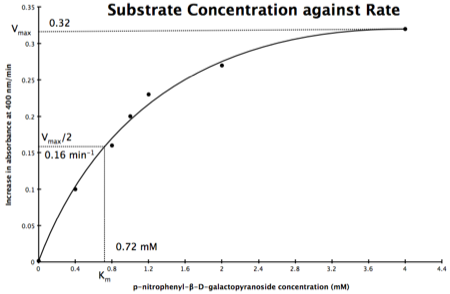
In the lab we can change the substrate concentration (S) in a reaction and measure the rate (v). As you know, a plot of substrate concentration (S) against rate (v; initial velocity) gives a curve, which plateaus at V
max, with K
m the concentration at half-V
max:

Plot of Substrate Concentration against Initial Velocity (rate)
The direct measurement for V
max can never be achieved in the lab as the concentration of the substrate needed would never be reached. Also, in the lab we would use a computer to calculate the values, and determine V
max, and K
m. However, you should be able to calculate V
max, and K
m yourself, just so you know the computer is right, and it is possible calculate these values from experimental data by using a linear plot and an equation derived from the Michaelis-Menton Equation.
As you know, the equation for a straight line is:

Equation for a straight line, where m = the gradient and c = the intercept on the y-axis
So, the problem is, how do we get:
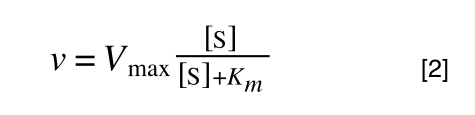
to look like equation 1 so we can plot a straight line using the terms we can measure, i.e v and S? The answer is, we rearrange....
The first thing we need to do is invert equation 2 to get:
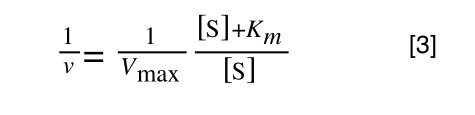
If you didn’t understand that ‘mathematical move’ consider this:
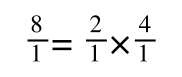
The above is true. That is, 2 over 1 = 2, 4 over 1 = 4 and 2 times 4 is 8. If I simply invert (flip) all the parts:

It is also true.
Equation 3 is getting closer to what we want as we now have 1/v and this is our y in equation 1. All we need to do now is ‘extract’ x (which is our substrate concentration) from equation 3.
So, we have:
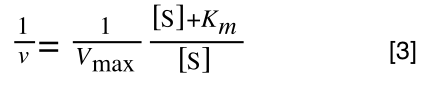
which is the same as:
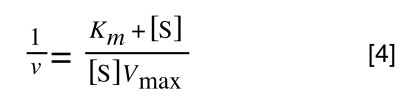
If you consider the following it is true:
we can ‘separate terms’ on this and express it in several other ways:
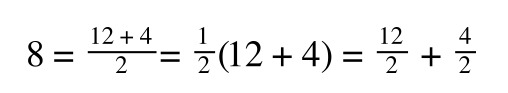
That is, once we find a ‘common’ element (in the above example 1/2, and in equation 5 1 over [S]V
max) we rearrange, so:

extracting 1 over [S]V
max in equation 4 we get:
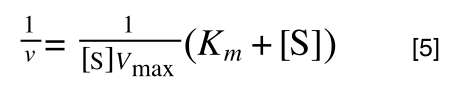
multiplying through with 1 over [S]V
max gives:

As you can see in equation 6 we have two terms after the + that can cancel out, and our experimental variable (S) can be separated, so:
/div>
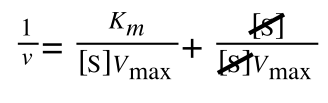
so we get:
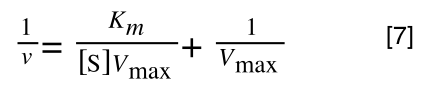
If that bit of maths has lost you, consider this:
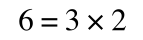
if you divide both sides by 2 it is still correct:
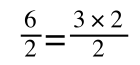
However, on the righthand side the two 2s can be cancelled to give:
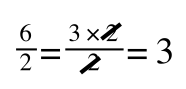
which is still correct.
Finally, separating out 1/[S] from 7 gives:
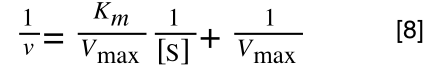
Which when compare to equation 1:

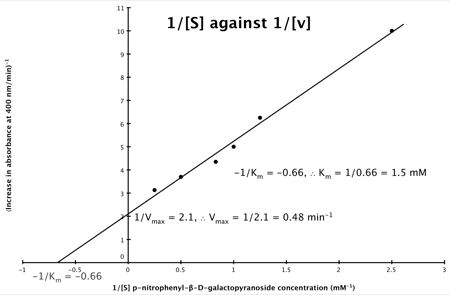
It can be seen that:
- y = 1/v
- x = 1/[S]
- c, the y intercept = 1/Vmax
- m, the gradient = Km/Vmax
And, the intercept for the x axis, (i.e. when rate (y) = 0) is 1/-K
m.
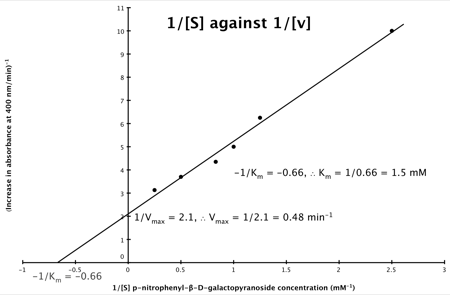
Hence, the final graph is:
If you would like to support my blogging efforts, then please feel free to buy me a coffee at https://www.buymeacoffee.com/drnickm
Additional Resources