I have been asked how you can track down papers online when a lecturer gives a paper reference as, for example:
Clin Med. (2003) 3, 333-7
The easiest way to track this down is to use 'Single Citation Matcher'.
Go to pubmed and the link can be found in the left-hand menu. Alternatively, follow this direct link:
http://www.ncbi.nlm.nih.gov/corehtml/query/static/citmatch.html
On the page enter the required information, so using the above reference, Clin Med. (2003) 3, 333-7, you should end up with:
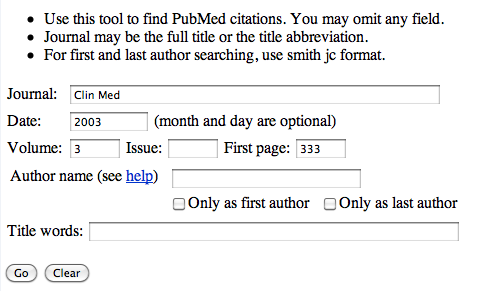
Click the 'Go' button and you should be taken to the paper (and any link to the full paper if available).
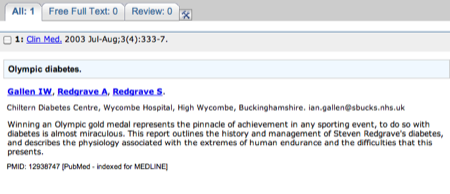
The abstract for the paper - don't forget there may be a link to the full paper on the far righthand side of the page.
If you would like to support my blogging efforts, then please feel free to buy me a coffee at https://www.buymeacoffee.com/drnickm