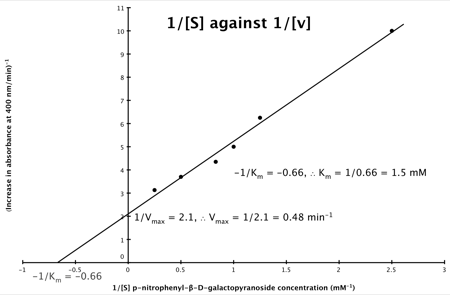
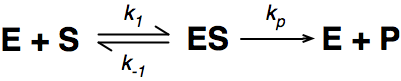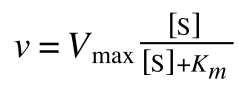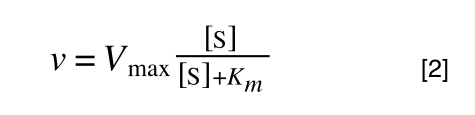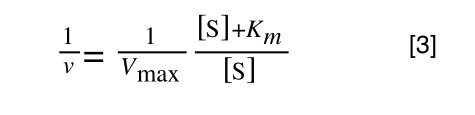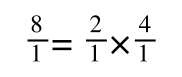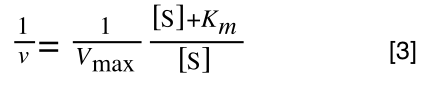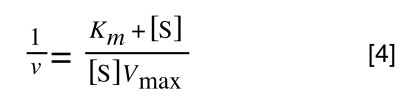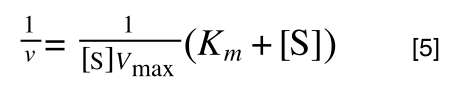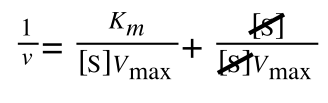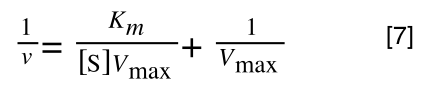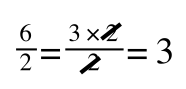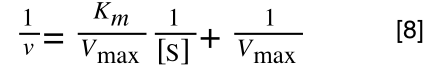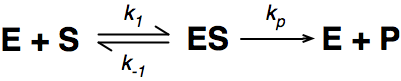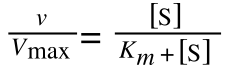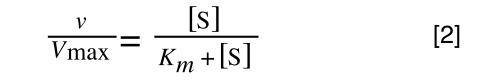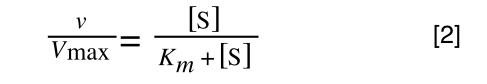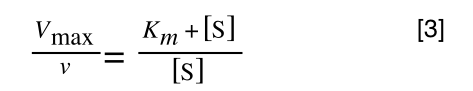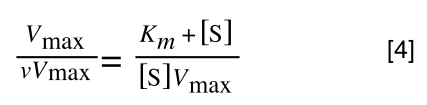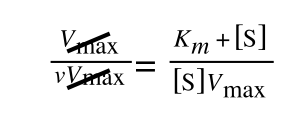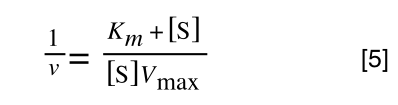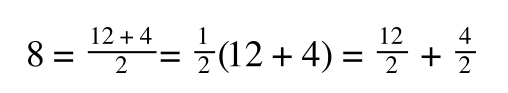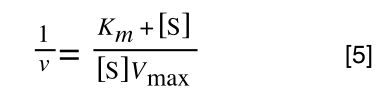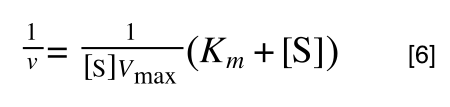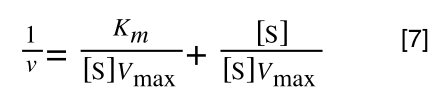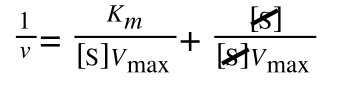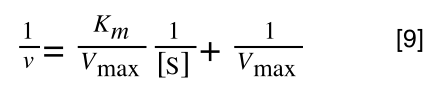A number of the class members have asked why (or how) they calculated the standard deviation incorrectly in the assessment.
Having looked at the class answers it would appear that the main reason students got it wrong is because they were calculating the standard deviation of a POPULATION and not of a SAMPLE.
The equation for the standard deviation of a population (remember a population is ALL possible examples of a measurement/object and is something that is normally difficult to measure - think of the apple example in the lecture - as all examples can't be collected) is:
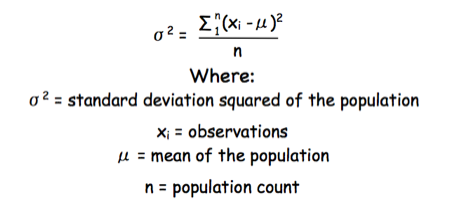
Equation for the standard deviation of a population
Whereas the standard deviation of a sample (which is what we had) is calculated by:

The equation for the standard deviation of a sample
Note that the real differences between the two equations are that in the calculation of the standard deviation of a population, we are using the true population mean (µ), whereas in the calculation of the standard deviation of a sample, we are using an estimate of the mean (x bar), plus in the calculation of the standard deviation of a sample we are using n - 1 (also called degrees of freedom) as this gives better estimate of the true standard deviation of a population (and we are working with a sample).
If you would like to support my blogging efforts, then please feel free to buy me a coffee at https://www.buymeacoffee.com/drnickm
Additional Resources
- 📗 - Maths and Chemistry Refresher for Life and Biomedical Scientists
- 📗 - Catchup Maths and Stats (affiliate link to the book)
- 📗 - Maths and Chemistry Refresher for Life and Biomedical Scientists
- 📗 - Catchup Maths and Stats (affiliate link to the book)